Co-Creating the Future of Beauty: SBX and POLA Chemical Industries Unveil AIM POLAR
Read the full press release from POLA CHEMICAL INDUSTRIES. here (in Japanese).
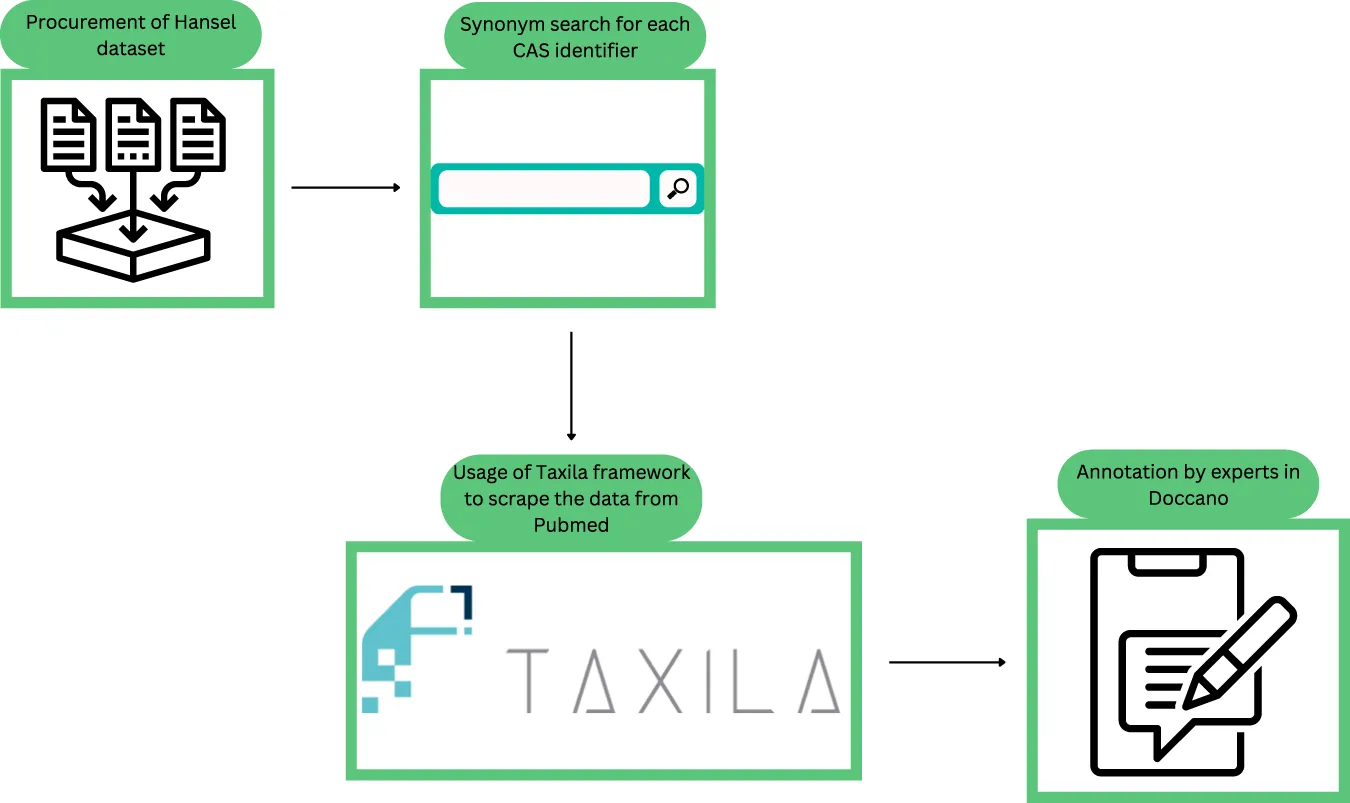
SBX is proud to announce our role in the launch of AIM POLAR, a revolutionary R&D platform unveiled by our partner, the cosmetics leader POLA CHEMICAL INDUSTRIES. The two companies jointly built this platform, combining POLA CHEMICAL INDUSTRIES deep industry knowledge with SBX’s technical expertise. In a true co-creation effort, SBX was responsible for the complete technical development of the system, from the powerful AI engine at its core to the intuitive user interface that brings its capabilities to life.
POLA CHEMICAL INDUSTRIES entrusted SBX with their vision for a transformative tool that could digitize their deep formulation expertise. Our team custom-built the backend AI engines to translate POLA CHEMICAL INDUSTRIES’s vast historical data into precise predictions for product texture, safety, and stability. In parallel, we designed and developed the intuitive touchpoint for POLA CHEMICAL INDUSTRIES’s researchers to have a seamless and powerful way to interact with the AI’s insights.